Alternative splicing alters the protein sequence of effector genes
Background
Alternative splicing (AS) is a molecular process that generates diverse mature mRNA molecules from single genes due to variations in intron splicing during transcription.
Although the biological significance of AS is well understood in plants and mammals, its role in fungi remains less explored. Research indicates that the frequency of AS varies significantly among different fungal species and is generally lower compared to other eukaryotes, ranging from less than 1% in the baker’s yeast Saccharomyces cerevisiae to 48.9% of the genes in the biocontrol fungus Trichoderma longibrachiatum. One hypothesis suggests that effector genes may undergo AS to modify their encoded protein sequences as a strategy to evade effector-triggered immunity in the host. To investigate this concept, I examined the AS landscape of the tomato pathogen Cladosporium fulvum.
Major findings
I found out that nearly 40% of the protein-coding genes of C. fulvum were predicted to be recurrently AS across the disparate infection timepoints.
The AS genes in C. fulvum showed preference for transporters, transcription factors, and cytochrome P450. Although most of candidate effector genes showed no evidence of AS, I dected AS in the previously described effector genes Ecp1, Ecp5, Ecp6, and Ecp12 that altered their encoded protein sequences.
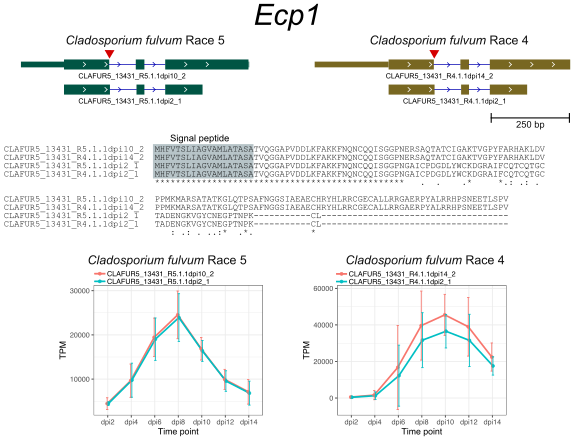
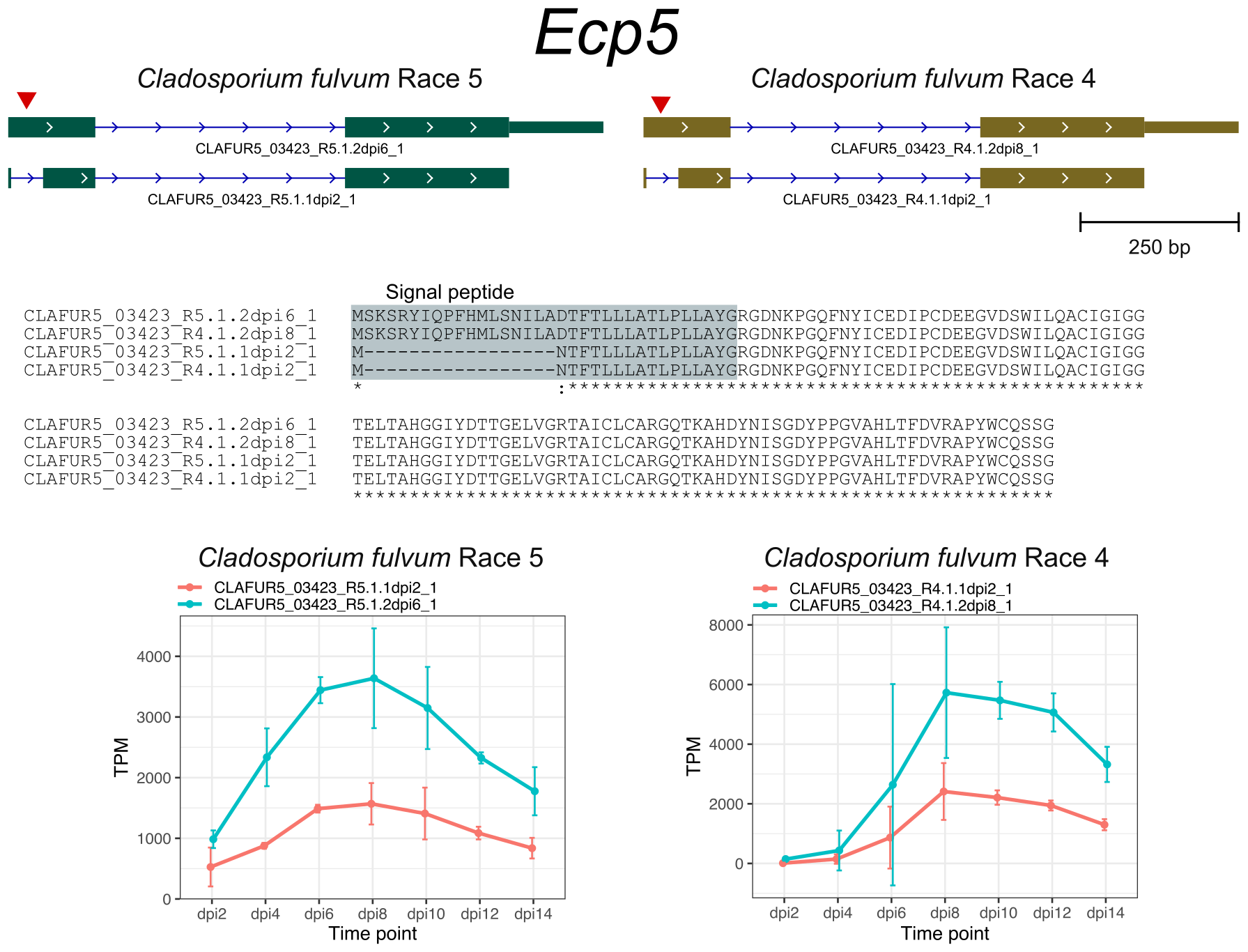
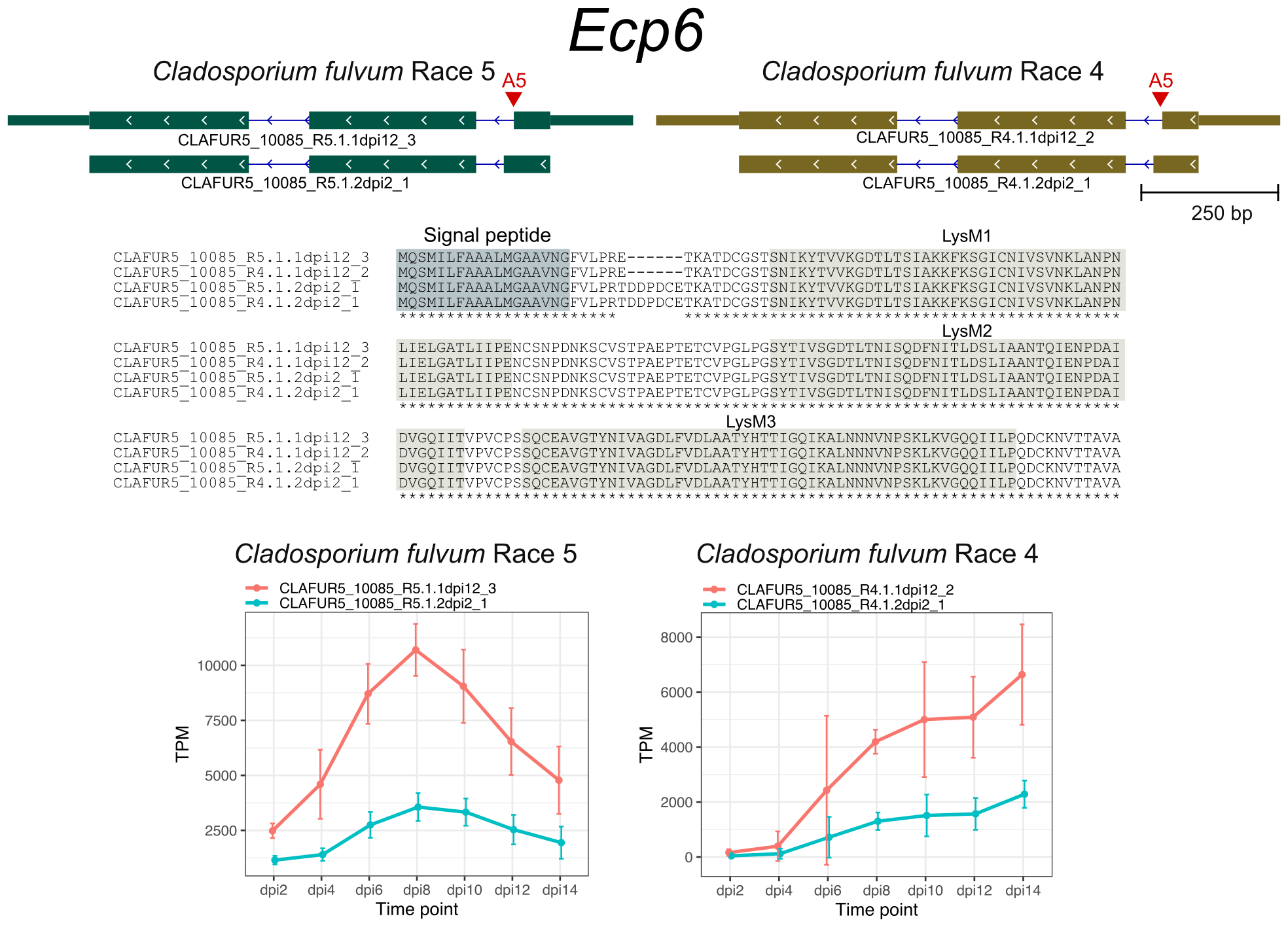
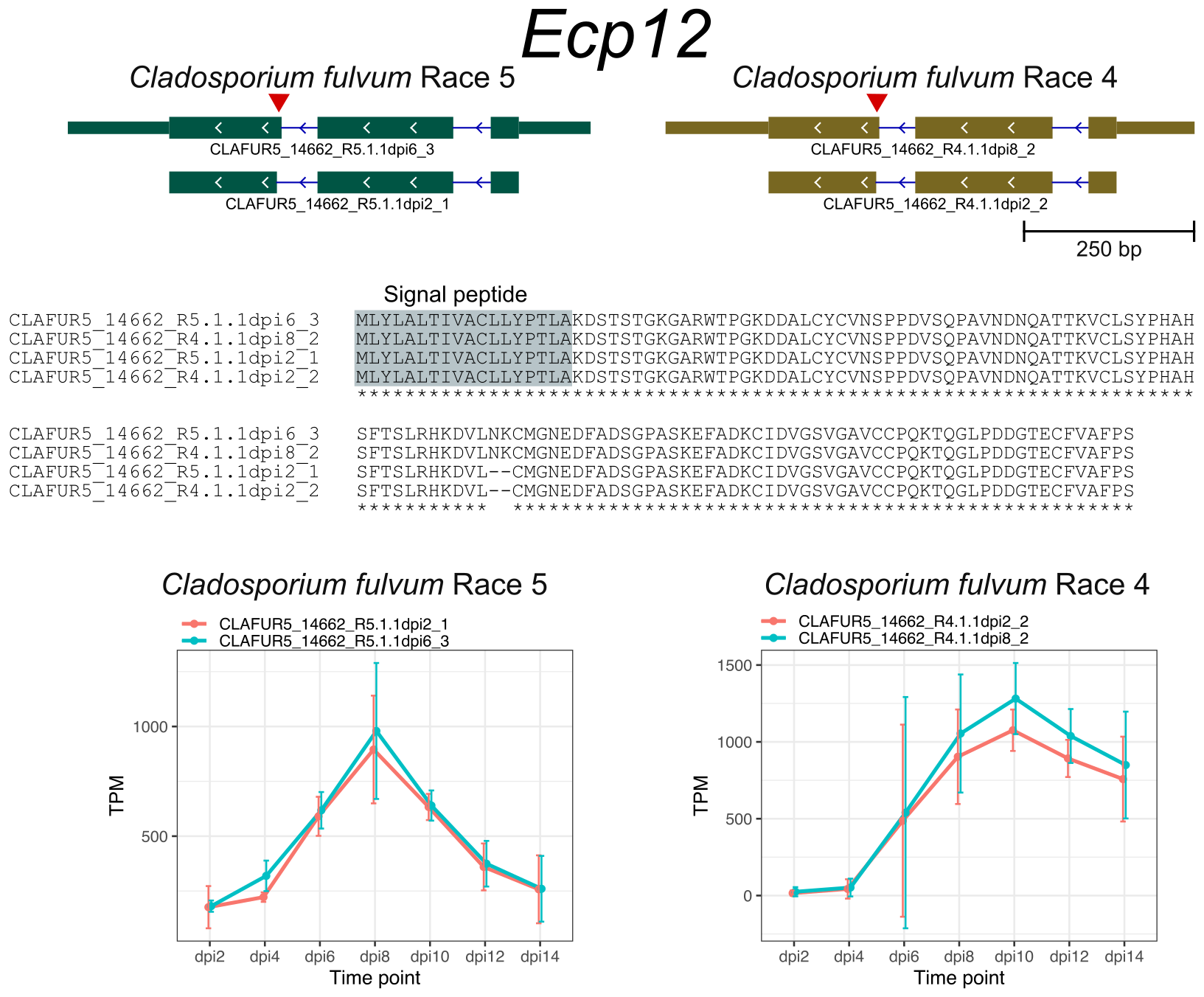
References
- Zaccaron, A. Z., Chen, L. H., & Stergiopoulos, I. (2024). Transcriptome analysis of two isolates of the tomato pathogen Cladosporium fulvum, uncovers genome-wide patterns of alternative splicing during a host infection cycle. PLoS Pathogens, 20(12), e1012791.
